In recombinant DNA technology, many different techniques are used to achieve different requirements or to obtain proper information so that an exact inference can be drawn out during genetic engineering. Gel electrophoresis, blotting techniques, bot-blot hybridisation, DNA sequencing, artificial gene synthesis, polymerase chain reaction, colony hybridisation, are some of the important techniques used.
Gel Electrophoresis
Gel electrophoresis involves separation of charged molecules (in aqueous phase) under the influence of an electrical field so that the molecules move on the gel towards the oppositely charged electrode, the cations move towards the negative electrode (anode) and anions move towards the positive electrode (cathode).
The genomic DNA is obtained from the desired host and then fragmented with restriction endonucleases. Gel electrophoresis is utilised for separating these cut fragments and isolating the desired DNA fragment. Agarose gel is most commonly used for separating the large segments of DNA, while the small DNA fragments (only a few base pairs long) are separated using polyacrylamide gel.
In gel electrophoresis, a buffer system is used which is a gel medium and a source of direct current. Samples having DNA fragments are applied on the gel and current is passed through for desired time period. Under the influence of the current applied, the DNA fragments move up to different distances on the gel on the basis of their charge to mass ratio.
The heavier DNA fragments travel a smaller distance, while the lighter ones travel a larger distance. After the migration of molecules, the gel is treated with certain stains so that the location of the separated molecules can be detected in the form of bands.
Blotting Techniques
Blotting is the transfer of macromolecules from the gel to the surface of an immobilising membrane. During this transfer, the bands of macromolecules on the membrane appear on the same positions as those on the gel. Nitrocellulose membrane, nylon membrane, carboxymethyl membrane, diazobenzyl-oxymethyl (DBM) membranes, etc. are the examples of some immobilising membranes which may be used in blotting.
Blot transfer technique is of three basic types, Southern blotting, Northern blotting, and Western blotting. Southern blotting is named after E.M. Southern (who developed this technique in 1975), and is used for transferring DNA from gel onto the membrane. Northern and Western blotting techniques are used for transferring RNA and protein bands, respectively. South-western blotting is another blotting technique which is used for examining the protein-DNA interactions.
Dot Blot Hybridisation
The procedure of dot blot hybridisation is nearly the same as blotting technique, with the only difference that the DNA fragments are separated by directly applying them as dots on the nitrocellulose membrane (instead of electrophoresis). Thereafter, the radioactively labelled DNA probes having the complementary base sequences to the desired DNA are applied on this membrane for its hybridisation. The position of this hybridisation is detected by autoradiography.
PCR (Polymerase Chain Reaction)
PCR technique discovered by Kary Mullis in 1985 is important in molecular biology (figure). It is employed for the amplification (or cloning) of a target DNA sequence. Sometimes it is also referred to as in vitro gene cloning (without expression of that gene) as it is carried out in vitro. Around billion copies of the target DNA sequence can be obtained from a single copy within few hours only using PCR.
PCR results in selective amplification of a selected DNA molecule. It has a limitation that the border region sequences of the DNA to be amplified should be known so that the appropriate primers can be selected to be annealed (attached) at its 3′ ends. Primer annealing is important as the DNA polymerase enzyme requires double stranded (ds) primer regions for initiating DNA synthesis.
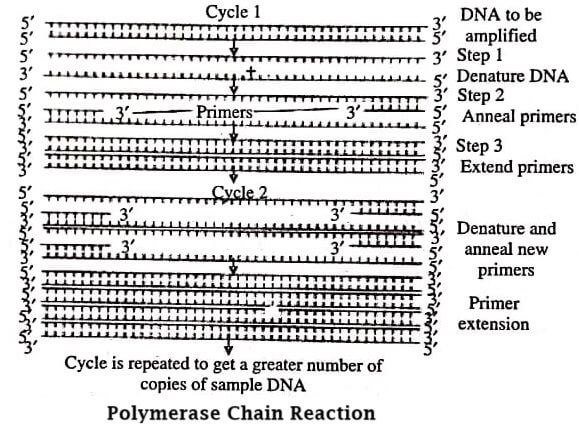
The PCR reaction takes place in eppendorf tube. The technique is used by the scientists in various disciplines since it is a quick, simple and extremely accurate technique. The major limitation of PCR is that being highly sensitive it may produce inaccurate results caused by several inhibitors or contaminating DNA segments that may be present in the sample DNA preparation.
Main Requirements of PCR
- Two nucleotide primers complementary to 3 ‘ ends of target DNA strands,
- Target DNA sequence,
- A heat stable DNA polymerase, e.g., Taq polymerase,
- Deoxy Adenosine Triphosphate (dATP),
- Deoxy Thymidine Triphosphate (dTTP),
- Deoxy Cytidine Triphosphate (dCTP),
- Deoxy Guanosine Triphosphate (dGTP), and
- A thermal cycler in which PCR is carried out.
Steps in PCR
- The target DNA sequence, excess of primers, dATP, dTTP, dCTP, dGTP, and Taq polymerase are mixed together in eppendorf tube, and the tube is placed in thermal cycler.
- The reaction mixture is provided with high temperature (90−98°C) for a few seconds for denaturing the DNA. Consequently, the double stranded DNA becomes single stranded.
- Temperature is lowered to 55°C for 20 seconds so that the primers are annealed at 3′ ends of DNA.
- Temperature is now maintained at 72°C for 30 seconds to facilitate the functioning of Taq polymerase and to synthesise the complementary strand of DNA.
- One cycle of PCR completes resulting in the formation of two ds DNA molecules from one ds DNA.
- The same cycle is repeated to obtain the required number of DNA copies.
Uses of PCR
- For DNA amplification,
- For detecting mutations,
- For diagnosing genetic disorders,
- For producing in vitro mutations,
- For preparing DNA for sequencing,
- For analysing genetic defects in single cells from human embryos,
- For identifying virus and bacteria in infectious diseases, and
- For characterisation of genotypes.
| Read More Topics |
| Expression of cloned gene within the host |
| Hybrid released translation (HRT) method |
| Vectors for cloning larger DNA fragments |





