The target DNA or the DNA insert isolated and enzymatically cleaved by the selective restriction endonuclease, are ligated (joined) through the action of ligase enzyme to the vector DNA. As a result, an rDNA molecule known as cloning-vector-insert DNA construct is produced.
Direct Ligation of Cohesive Termini (DNA Ligase)
The target gene can be inserted into a vector when both are cleaved with the same restriction enzyme, producing identical cohesive ends. A staggered two stranded cut ends complementary to each other in sequence, are produced (figure).
When performed annealing, the joint linking two DNAs bear one pair of staggered nicks on each strand. The two DNAs are connected with DNA ligase under suitable conditions to create a phosphodiester linkage among the adjacent nucleotide.
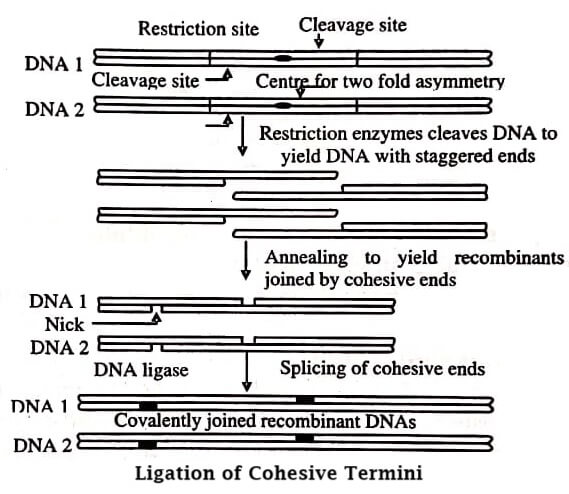
Generally, ligation is performed for 12−24 hours at 4−15°C temperature and is catalysed by either T4 DNA ligase or E. coli DNA ligase. The former uses ATP as a cofactor, while the latter uses NAD+. The ligated fragments thus produced have a mixture of DNA fragments and linear vector, undesirable circularised vector with no inserts, length of DNA formed from two or more fragments, circularised fragments, two or more vector molecules connected together, etc.
DNA Ligase
DNA ligase (or molecular glue) forms phosphodiester bond to join two DNA fragments. They repair the single stranded nicks in DNA double helix. In rDNA technology, they seal the nicks between adjacent nucleotides.
Blunt End Ligation
It is possible to transform any cohesive ends into blunt ends, by eliminating the protruding nucleotides using SI nuclease that will break the single stranded protruding DNA. The cohesive ends can also be transformed by filling in the single stranded ends by DNA polymerase. The blunt end ligation has a disadvantage that once ligated (using T4 DNA ligase), the DNA cannot be detached and becomes less efficient.
Linkers and Adapters
The blunt end molecules can be ligated through single strands of synthetic oligonucleotides with previously formed restriction sequences. The ligation conditions remain the same as required by blunt ended ligation. Upon annealing to each other these single stranded linkers become ligated to the DNA ends. After ligation each end of DNA molecules carries several linkers. When treated with restriction endonuclease, the pieces of restricted linker are separated, and the linker terminated DNA is ready for ligation.
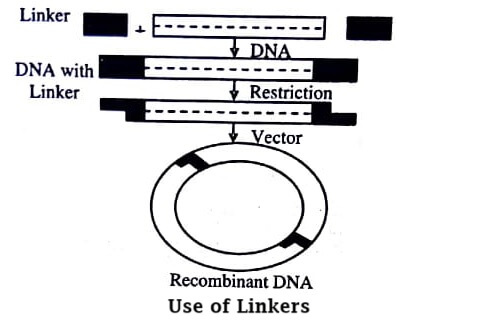
Adapters
Linkers and adapters resolve the problem of compatibility. Adapters are synthetic oligonucleotides of double stranded molecules. They have a blunt end on one side and a cohesive single strand tail on the other side.
Homopolymer Tailing
Oligonucleotide is attached to the end of a DNA strand through terminal deoxynucleotidyl transferase in order to join the blunt ended molecules. This forms intermolecular bonds between the vector and insert. The reaction of terminal transferase adds a tail to blunt ended DNA with protruding 3-hydroxyl terminus.
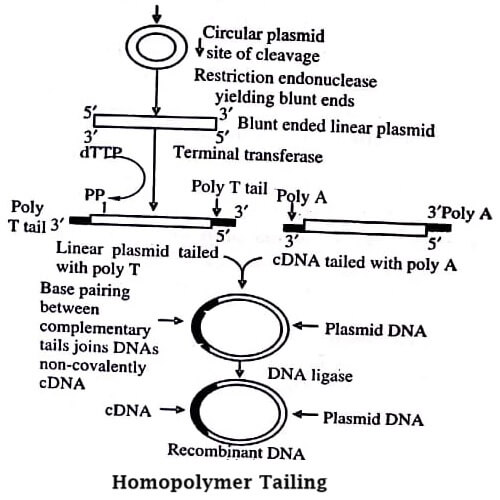
Vector and inserts are separately treated with terminal transferase and dATP or TTP (Figure). This builds up poly dA tails on one 3′ -termini and poly T tails on the other. When these complementary tails are mixes, stable, hybrid molecules that can be for transformation are produced. In the absence of ligation, the homopolymer tails result in a stable recombinant.
| Read More Topics |
| Introduction for recombinant DNA technology |
| Genetic engineering |
| Classification of fatty acids |